Plant-and resource modules
Design of plant-and resource benchmarks
Benchmarks concerning the plant and resource modules are clustered in sets belonging to a certain plant growth concept (i.e., plant module). Each set has a reference plant, which is an individual plant, growing with the particular growth concept but without competition or resource limitation (Figure 1a). The output metrics of each benchmark are the plant geometry parameters available for the particular plant module, e.g., stem height, crown, root and stem radius (Figure 1b). Additional outputs relevant for the tested module might be specified.
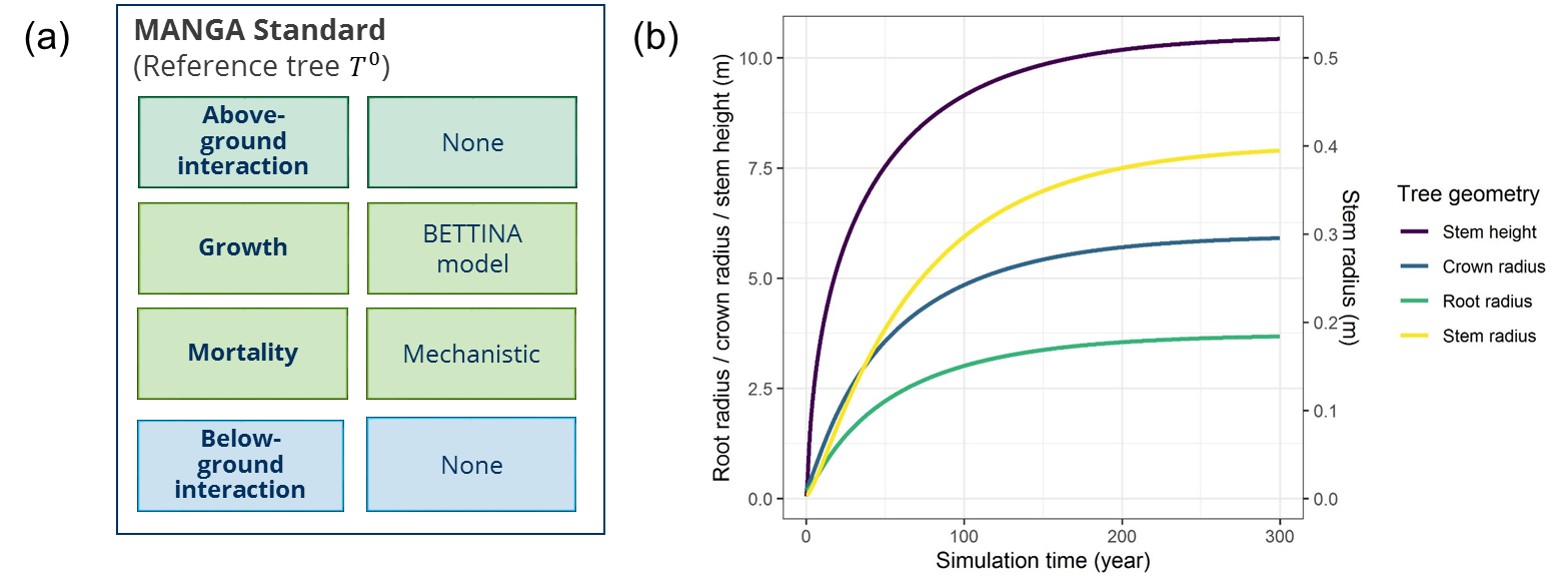
Figure 1: (a) Module combination to create the pyMANGA reference plant (T0). (b) Geometry of T0 over time.
Within a set, benchmarks are classified based on the compartment of the module tested, i.e. below-ground interaction, above-ground interaction and plant mortality. Modules of the other compartments are defined as for the reference plant (Figure 2a). This means, for example, to test the below-ground module ‘FixedSalinity’, above-ground interaction is disabled (SimpleTest) and mortality is mechanistic.
The general structure of each benchmark is a 2-plant setup, without recruitment (Figure 2b). The plants are placed centered in a 22x22 m model domain with a fixed distance of 2 m. The initialization of those plants is based on the geometry of the reference plant. The initial population is designed in a way that the respective geometry triggers a potentially critical situation of the concept. In some below-ground modules, this is for example the case, when the root systems of the plants overlap. Thus, we defined an initial population for each compartment. Each initial population is stored in ’Benchmarks/ModuleBenchmarks/PlantModules/<plant_module>/<compartment>/’.
If land- and seaward boundary conditions need to be defined in a setup, salinity is set to 25 ppt at the landward boundary and to 35 ppt at the seaward boundary. Random seed of all setups is 643879.
Time step length is 1e6 seconds (~12 days) and simulation time is 5e8 secs (~ 15.8 years, i.e. 500 time steps). Output is written after the first and last time step. The first output is used for automatic testing.
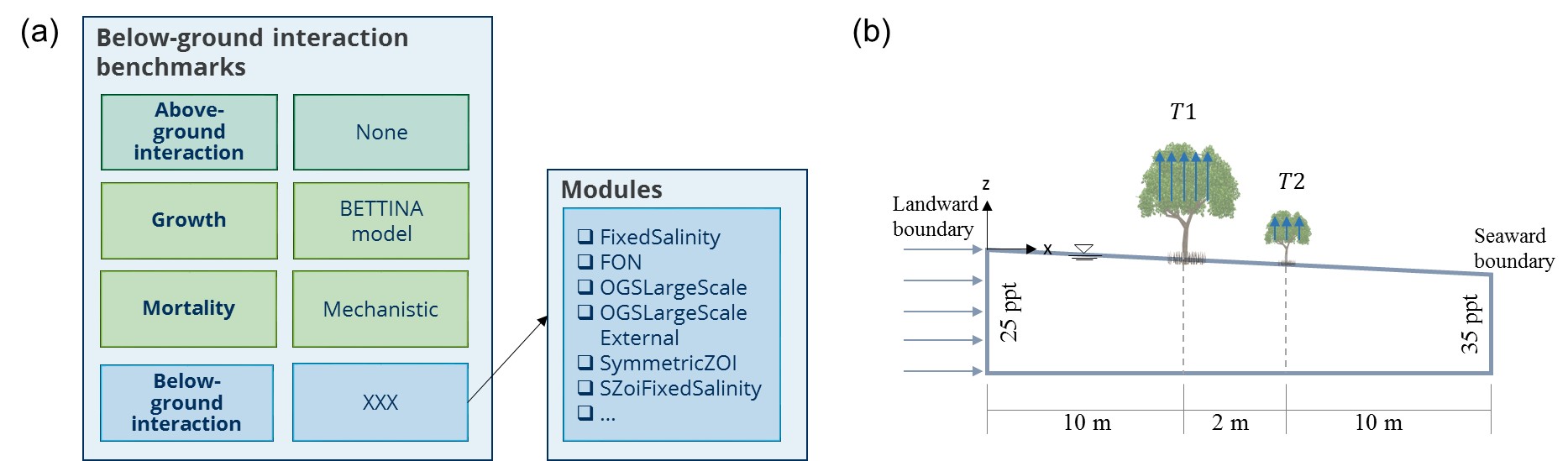
Figure 2: (a) Overview below-ground interaction setups. (b) Schematic representation of benchmark setup (based on Bathmann et al. 2020).
The following files must be provided for each benchmark
- pyMANGA project file
- Results csv file (
<output_times> [2e6, 5e8] </output_times>) of typeOneTimestepOneFile - All files required to run the setup, e.g. OGS project file (if applicable)
The files are stored in ‘Benchmarks/ModuleBenchmarks/PlantModules/<plant_module>/<compartment>/’ with the name of the module tested.
Overview of existing modules and benchmarks
Below is an overview of which modules and benchmarks are available. Modules marked in blue have already been benchmarked. Click on the button to go to the relevant folder.
Modules tested with Bettina
Bettina works with all resource and mortality modules and has therefore the most comprehensive collection of benchmarks.
Mortality
NoGrowth Memory RandomGrowth Random
Parameter 'k_die' set extremely low in order to force trees to die
Modules tested with BettinaNetwork
Network modules enable the formation of groups, representing root grafted trees. Grafted trees can exchange water, based on the water potential gradient between them.
The module describing the group formation as well as the water exchange is ‘SimpleNetwork’. All other below-ground network modules are children of this module and one or two other existing below-ground modules allowing for example the definition of a salinity gradient (‘NetworkFixedSalinity’). Network below-ground modules must be combined with the growth module ‘BettinaNetwork’ which is based on ‘Bettina’ but describes resource allocation to form the root graft.
Mortality
NoGrowth Memory RandomGrowth Random
Parameter 'k_die' set extremely low in order to force trees to die
Modules tested with Kiwi
In Kiwi, crown and root radius are not explicitly defined as in Bettina as the tree is only described by dbh and height. Hence, below- and above-ground modules, which depend on crown and root radius would not work. We therefore use the equation suggested by Berger and Hildenbrandt (2000) to scale dbh to the so called zone of influence to estimate root and crown radii. This means however that root and crown radii are equal.
Mortality
NoGrowth Memory RandomGrowth Random
Parameter 'k_die' set extremely low in order to force trees to die

